Human Serum Albumin (HSA) as a diabetic biomarker



Collaborators:
Prof Shinji Saito (IMS, Japan)
Asst Prof Toshifumi Mori (IMS, Japan)
Dr Deanpen Japrung (NANOTEC, THA)
Human serum albumin (HSA) is the most abundant transport protein in blood. It has astonishing capacities to bind metabolites and a wide range of drugs (i.e. warfarin, ibuprofen, and indomethacin). Physiological and pathological changes can induce an alteration of protein conformation and efficiency of ligand binding resulting in the function impairment in HSA. Covalently binding to glucose (glycation) is one of basic impairments found in HSA. Glycated HSA (GHSA) has been used as a biomarker for glycemic control. Especially, earlier studies observed high concentration of GHSA in diabetic patients. Thus, many studies were devoted to explore the feasibility of GHSA as a diabetes biomarker.
In addition to glucose, galactose and fructose were also able to bind to HSA. Glucose and galactose are hexose monosaccharide where galactose is a C4 epimer of glucose. Both sugars were found to share several glycation sites and similar glycation mechanism. Apart from diabetes with glucated HSA, monitoring galactated proteins may provide a useful tool for galactosemia control. Some other monosaccharides and carbohydrates can also bind to HSA.
Our group employs Molecular Dynamics (MD) simulations to understand the binding mechanism and affinity of sugars. Our first aim is to understand the nature of sugar binding activities and reveal how such binding impacts on protein conformation and function. When such processes are well understood, we then explore the possibility of glycated HSA as a diabetic biomarker. The designed aptamer has been used to detect the level of GHSA in a sample. Also, our collaborator uses this aptamer to design an aptasensor. Our second task is to explain how this aptamer binds and detect only GHSA in order to improve the aptamer selectivity. There are still a lot of interesting work to do.
Publications:
1. Pongprayoon P and Gleeson MP*. Probing the Binding Site Characteristics of Human Serum Albumin: A Combined Molecular Dynamics and Cheminformatics Investigation, J Mol Graph Model, 2014, 54:164-173
2. Awang T, Wiriyatanakorn, N, Saparpakorn P, Japrung D, and Pongprayoon P*, Understanding the effects of two bound glucose in Sudlow site I on structure and function of human serum albumin: Theoretical studies, J Bio Struc Dyn, 2016, 29: 1-10
3. Chayachon A, Luksirikul P, Kankla P. Pongprayoon P, Tarattrakoon, K, Paiboonsukwong K, Fuchareon, S, Dharakul T, and Japrung D*. Graphene based Aptasensor for Glycated Albumin in Diabetes Mellitus Diagnosis and Monitoring, Biosens Bioelectron, 2016, 82:140-145
4. Panman W, Japrung D, and Pongprayoon P*, Exploring the interactions of aptamer with human serum albumin: Simulation studies, J Bio Struc Dyn, 2016, 1-25
5. Pongprayoon, P* and Mori, T*. Critical role of dimer formation in monosaccharide binding to human serum albumin, PCCP (2017).
Outer Membrane Proteins (OMPs) and their transport properties

Collaborators:
Prof Syma Khalid (U. of Southampton, UK)
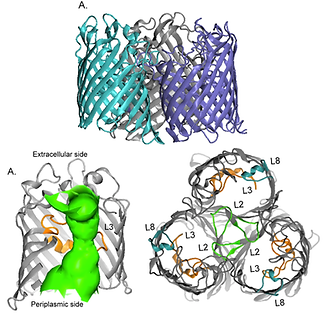
Gram-negative bacteria are a leading human pathogen. The major bottleneck to fight against these bacterial is their impermeable outer membrane (OM). Their membrane contains protein channels, called outer membrane protein (OMP) which facilitate nutrient and ion transport. In advanced bacteria, only substrate-specific OMPs are available therefore they becomes very difficult to treat. Also, such bacteria become drug resistance very quickly. The understanding on strucutre and function of OMPs are crucial. Especially, the clear mechanism of drug penetration through such membrane channels plays a key role in treatment of bacterial infection. Many studies revealed that some OMPs serve as a drug entryway.
Apart from structure and function, we're also interested in drug translocation through OMPs. We perform MD simulations to better understand mechanisms of nutrient, ion, and drug transport in atomic level.
Publications:
1. Pongprayoon P, Beckstein O, Wee CL, and Sansom MSP*. Simulations of anion transport through OprP reveal the molecular basis for high affinity and selectivity for phosphate, PNAS. 2009, 106:21614-21618.
2. Pongprayoon P*. How do the protonation states of E296 and D312 in OmpF and D299 and D315 in homologous OmpC affect protein structure and dynamics? Simulation studies, Comp Bio Chem, 2014, 53:226-234
3. Niramitranon J, Sansom MSP, and Pongprayoon P*. Why do the outer membrane proteins OmpF from E.coli and OprP from P.aeruginosa prefer trimers? Simulation studies, J Mol Graph Model, 2016, 10:1-7
4.Soomboon, K, Niramitranon J, and Pongprayoon P*. Probing the binding affinities of imipenem and ertapenem for outer membrane carboxylate channel D1 (OccD1) from P. aeruginosa: simulation studies. J Mol Mod, accepted.
Betaine Aldehyde Dehydrogenase (BADH2) in rice


Collaborator:
Asst Prof Ranjit Vijayan (Department of Biology, UAE University, UAE)
Betaine aldehyde dehydrogenase 2 (BADH2) is an enzyme that inhibits the accumulation of 2-acetyl-1-pyrroline (2AP), a potent flavor compound in rice fragrance. BADH2 contains 3 domains (NAD-binding, substrate-binding, and oligomerization domains). It catalyzes the oxidation of amino aldehydes. The lack of BADH2 results in the formation of 2AP and consequently an increase in rice fragrance. To date, inadequate data on BADH2 structure and function are available. An insight into the nature of BADH2 can serve as one of key starting points for the production of high quality fragrant rice.
Here, we employed Molecular Dynamics simulations (MD) to primarily understand the structure and function of BADH2.
Publications:
1.Baichareon, A, Vijayan, R, and Pongprayoon, P*. Structural insights into betaine aldehyde dehydrogenase (BADH2) from Oryza sativa explored by modeling and simulations. Sci Rep, 2018.
Human Defensin 5 (HD5) as antimicrobial peptides


Human Defensin 5 (HD5) is a host-defense peptide that is expressed in Paneth cells. HD5 displays the antimicrobial properties. It also has chemokine-like activities. They can act as chemotactic attractants for human monocytes and dendritic cells and T cells. To date, there are many studies indicate different forms of HD5 oligomers.
Here, we are interested in structure and function of HD5. We focus on their antimicrobial mechanisms. We want to explore how such HD5 can kill microbes. We also have a collaborator in the hospital to help out.
Collaborators:
Prof Shinji Saito (IMS, Japan)
Dr Phoom Chairattana (Siriraj hospital, Bangkok, THA)
Publications:
1.Awang, T and Pongprayoon, P*. The Adsorption of human defensin 5 on bacterial membranes: simulation studies. JMMO, 2018.
